1. The main defender of dolphins is Rick O'Barry
3. Rick O'Barry became involved with trying to save dolphins because in his opinion, he contributed to their popularity which led to their cruel treatment.
5. Dolphins are in the mammalian order of cetacean.
7. The small islands in the Caribbean is supporting whaling because the Japanese is paying them to support whaling.
9. Some dolphins are stabbed to slow them down.
11. The main point of this movie was to inform people about the horrible Japanese practice and the issues with eating dolphin meat.
13. Biomagnification occurs when the concentration of the toxic substance is increased all the way to the food chain.
15. The fishermen refuse to stop despite being offered payment because they see killing dolphins as pest control. They think dolphins eat all the fish in the ocean and contribute to their declining numbers.
17. They disguise cameras by making them look like the rocks found near the cove.
19. When the Deputy of Fisheries claim that the dolphins were killed humanely, one of the crew members showed the Deputy the secret footage of the cruel slaughtering of the dolphins.
21. When the two divers first saw the cove, they witnessed a struggling bleeding dolphin taking its last breath.
Saturday, 6 June 2015
Photosynthesis vs Cellular Respiration
Photosynthesis: Synthesizing molecules from smaller components in order to store energy, requires energy in the process.
- Noncyclic Photophosphorylation
Cellular Respiration: breaking down complex molecules into smaller units, releasing energy.
- Glycolysis:
- Noncyclic Photophosphorylation
- P680 absorbs a photon, the excited electron goes to b6-f complex, then to P700 where electrons get excited by NADP reductase to reduce NAP+.
- Oxygen and NADPH are formed
- can produce ATP
- electron in P700 is excited by a photon, instead of being used to reduce NADP+ like in noncyclic photophosphorylation, electrons from Fd is passed to b6-f complex and back to P700.
- no NADPH or oxygen if formed
- can produce more ATP the little NADP is available.
- rubisco fixs CO2 and RuBP(5C) to become 2 PGA(3C) ---> 2 BPG(3C) ---> 2 G3P(3C)
- some G3P are used to make glucose, others recycled(G3Px2---> 1 glucose)
- uses ATP and NADPH
Cellular Respiration: breaking down complex molecules into smaller units, releasing energy.
- Glycolysis:
- Glucose---> Glucose 6-phosphate ---> Fructose 6-phosphate ---> Fructose 1,6-phosphate ---> G3P---> 2 BPG ---> 3PG---> 2PG ---> 2 PEP --->2 pyruvate
- 2 ATP consumes, 4 ATP produced, 2 H2O produced
- OXAL(4C) +Acetyl-CoA(2C) --->CIT(6C) ---> ISO(6C) --->alphaKG(5C) ---> Succinyl-CoA(4C) --->SUC(4C) ---> FUM(4C) ---> MAL(4C) ---> OXAL(4C)
- produces 2 ATP, 6 NADH, 2 FADH2
- 2H+ ---> 1 ATP
- NADH passes 2 electrons to NADH dehydrogenase ---> bc1 complex ---> Cytochrome oxidase complex => 6 H+ 1 NADP ---> 3 ATP, 24 ATP(2 NADP from glycolysis behaves like FADH2)
- FADH2 passes 2 electrons to bc1 complex ---> Cytochrome oxidase complex => 4H+ 1 FADH2 ---> 2 ATP, 8 ATP
Monday, 18 May 2015
Photosynthesis- Light Dependent Rnx vs Light Independent Rnx
Photosynthesis:
- 6CO2 + 6H2O --light---> C6H12O6 + 6O2
- plants to produce food
- make up of two reactions: light dependent and light independent rnx
Light Dependent Reaction:
- takes place in the chloroplasts of cells.
- two membranes, and contains several stacks of thylakoids within the stroma on the inner membrane.
- uses sun's energy to remove electrons from water, transferring them from proteins through redox rnx, giving rise to the reduction of NADP+ to NADPH
- electron transport chain (ETC):
- 6CO2 + 6H2O --light---> C6H12O6 + 6O2
- plants to produce food
- make up of two reactions: light dependent and light independent rnx
Light Dependent Reaction:
- takes place in the chloroplasts of cells.
- two membranes, and contains several stacks of thylakoids within the stroma on the inner membrane.
- uses sun's energy to remove electrons from water, transferring them from proteins through redox rnx, giving rise to the reduction of NADP+ to NADPH
- electron transport chain (ETC):
- PSII and PSI both need to absorb light in order to excite the electron, which is gained from water, and transport it.
- The pathway of the electron throughout the process: water---> PSII---> PQ --->b6f ---> PSI---> Fd---> FNR ---> NADP
- Two hydrogen ions were released when the water break down outside the PSII. Hydrogen ions also enter PQ to form PQH2. To from the neutral NADPH, hydrogen ion enters to play the role to balance the electron. Large amount of hydrogen ions go through the ATP synthase to slow down its spinning motion in order to form ATP.
- The process of breaking down molecules by light is called photolysis.
- Gaining electron of a molecule is called oxidation; losing electron of a molecule is called reduction.
- Chemiosmosis is the movement of chemicals from high concentration to low concentration through a selectively permeable membrane. The process of releasing ATP is chemiosmosis.
- The one help to make ATP is called ATP synthase.
- It is important that the oxygen leave the water molecule at the beginning, because it will come back again.
- ETC happens in thylakoid lumen.
- When the spinning motion of ATP synthase slows down by hydrogen, a phosphate come to take away the ADP which has two phosphate atoms in it and from ATP which has three phosphate atoms in it.
- assimilates carbon dioxide to produce an organic molecule that can be used to produce biologically important molecules such as carbohydrates.
- energy from ATP and NADPH is used to synthesize glucose.
Pig Dissection
In this dissection, we mainly looked at three systems: the nervous system, the endocrine system and the urinary system.
First day: dissection of the lower body
We examined the pig's gender first. We believed it was a male, which was proved by the two testes we found.
The eye ball and lens
First day: dissection of the lower body
Then we isolated the liver.
The stomach & intestine
We opened the stomach.
The pancreas
The kidneys
Second day: dissection of the upper body
The first organ we isolated were the lungs
The heart
The brain was extremely soft. Unfortunately we didn't isolate the brain stem.
The eye ball and lens
Sunday, 5 April 2015
Neurons
Neurons:
- have a cell membrane, cytoplasm, mitochondria, and a nucleus.
- have specialized cell structure that enable them to transmit nerve impulses.
- different shapes and sizes.
- common features: dendrites, a cell body, am axon, and branching ends.
- Dendrites:
- short, branching terminals that receive nerve impulses from other neurons or sensory receptors, and relay the impulse to the cell body.
- numerous and highly branched: increases the surface area
- Cell body:
- contains the nucleus and is the site of the cell's metabolic reactions.
- processes input from the dendrites.
- Axon:
- conducts impulses away from the cell body.
- ranges from 1mm-1m.
- In order to communicate with the nearby neurons, glands or muscles, axon terminal releases chemical signals into the space between it and the receptors or dendrites.
- Myelin Sheath:
- fatty insulating layer of some neurons.
- gives the axons white appearance.
- protects neurons and speeds the rate of nerve impulse transmission.
- Schwann Cells:
glial cell, from myelin by wrapping themselves around axon.
Types of Neurons (Classified Structurally)
Types of Neurons (Classified Functionally)
Reflect Arc:
- Moves directly to and from brain or spinal cord before the brain centres involved with voluntary control have time to process the sensory information.
- Reflex arcs:
- Simple connections of neurons that result in reflexive behaviours.
- Reflexes:
Sunday, 29 March 2015
Sunday, 1 March 2015
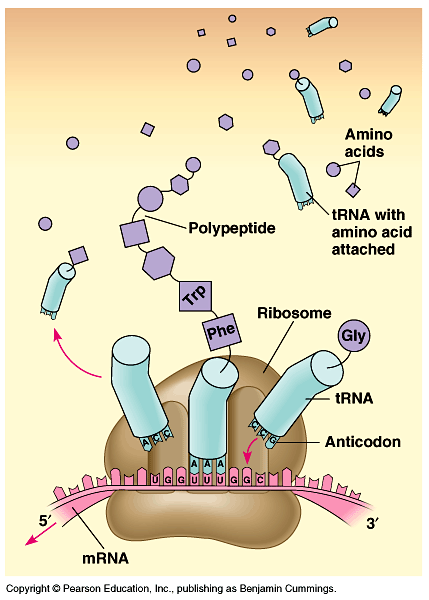
Translation
Act I: Initiation
Act II: Elongation
Act III: Termination
- The mRNA comes to the cytoplasm, with a starting codon AUG.
- A small ribosomal subunit binds to mRNA. The ribosome has two ribosomal units (small and large).
- The first tRNA carrying a specific amino acid, met, at one end and having a specific nucleotide triplet, anticodon, UAC, at the other end comes to bind to the starting codon.
- The ribosome also has one site for binding of mRNA and three sites for the binding of tRNA (P site, A site, E site).
- A site: new tRNA with next amino acid to be added to the chain.
- P site: holds growing polypeptide chain.
- E site: discharged tRNA which will return to cytoplasm and pick up designated amino acid.
- Initiation factors brings the large ribosomal subunit to mRNA, placing the tRNA in the P site.
Act II: Elongation
- Another tRNA carrying amino acid recognizes its corresponding codon at the A site.
- An RNA molecule catalyzes the formation of a peptide bond between the polypeptide in the P site with the new amino acid in the A site.
- The polypeptide chain is transferred to the tRNA at the A site.
- The ribosome moves the tRNA with the attached polypeptide from the A site to the P site. This process need energy provided by GTP.
- The first tRNA enters the E site, and as the third tRNA attaches to the A site, it exits the E site to the cytoplasm to pick another amino acid.
Act III: Termination
- When one of the three stop codons UAG, UAA, UGA reaches the A site, a release factor cut the bond between the polypeptide chain and its tRNA at the P site.
- Polypeptide, which is known as protein, is released.
- Translation complex disassembles.
Transcription
Transcription is the copying of a sequence of DNA to produce a complementary strand of RNA
Act I: Initiation
Act I: Initiation
- Transcription factors recognize promoter regions (TATA box) on the template and bind to the promoters.
- This signals RNA polymerase II to bind to transcription factors. Together they form a transcription initiation complex.
- Polymerase II starts transcription.
Act II: Elongation
- RNA polymerase unwinds DNA double helix and adds RNA nucleotides to the 3' end of the growing strand. RNA, same as DNA, grows from 5' to 3'.
- When adding the complimentary nucleotides to the RNA transcript, every thymine is replaced by uracil.
- As RNA polymerase II moves forward, the double helix behind re-forms, and the newly transcribed RNA molecule, RNA transcript, peels away.
- The strand that RNA reads is called the template strand/ antisense strand.
- The strand that has exactly same sequence as RNA is called coding strand/ sense strand.
- A single gene can be transcribed by multiple polymerase simultaneously.
Act III: Termination
- Transcription stops when RNA polymerase transcribes as a terminator AAUAA.
- The pre-mRNA is cut, and RNA polymerase II is released form the DNA.
- In eukaryotic cells, before the mRNA enter the cytoplasm, enzymes work on modifying them.
- G-cap (modified guanine) is assed to the 5' end of pre-mRNA.
- PolyA-tail is added to the 3' end of pre-mRNA.
- Pre-mRNA combines with snRNPs and other proteins to form a spliceosome.
- The snRNA in the snRNPs base pair with nucleotides at the end of the noncoding segments on the pre-mRNA, introns.
- Introns are excised from the pre-mRNA, then the coding regions, exons, are spliced together in the spliceosome.
- The snRNA acts as a ribozyme, and the RNA acts as an enzyme.
- The spliceosome comes apart, releasing mRNA.
Sunday, 22 February 2015
DNA Replication
Act I: Initiation
- Helicase unwinds the double-stranded DNA at locations called replication origins. The structure that is created is known as "replication fork".
- Single-strand-binding-proteins then help to stabilize the newly unwound single strand, and prevent it from rebinding with its other strand.
- As the DNA unwinds, tension starts to build up on either sides of the unwound strand. Gyrase then comes in and cuts the two ends to release the tension.
- An RNA primer is added on to the DNA template strands by the enzyme primase to initiate the DNA replication process.
Act II: Elongation
- DNA polymerase III starts adding new nucleotides to the end of the RNA primers to create a new DNA daughter strand complementary to the original strand.
- SInce DNA is antiparallel, the new strands will grow in the opposite direction of each other.
- Leading strand: synthesized continuously in the 5'->3' direction by polymerase III. Only one RNA primer is needed to initiate the replication.
- Lagging strand: synthesized discontinuously in the 3'->5' direction by polymerase III. In the lagging strand, the RNA primase adds many RNA primers to form short, discontinuous fragments known as Okazaki fragments.
Act III: Termination
- Polymerase I comes in and now proofreads the lagging strands and replaces the RNA primers with DNA nucleotides.
- As a polymerase III reaches a previous RNA coded section, the Okazaki segments are glues together with ligase.
'
Subscribe to:
Comments (Atom)